My PhD: Part 2: the ichthyosaurs strike back
In the last post, I talked about the British Late Jurassic ichthyosaurs. This will make up the first part of my thesis. The other main part will look at ichthyosaur phylogeny. The last part will be about further projects that arise from these.
- Part 1: a new ichthyosaur
- Part 2: the ichthyosaurs strike back
- Part 3: the return of the ichthyosaurs
Summary
- Three phylogenetic analyses were published in 1999–2000 (Motani 1999; Maisch and Matzke 2000; Sander 2000).
- Though many parts are similar, there are many differences in data and results.</li>
- I will carry out an amalgamative analysis of these three studies in a similar fashion to Ketchum and Benson’s (2010) study on the Plesiosauria.
First things first: what is phylogeny?
Phylogeny
Phylogenetics is the study of the relationships between organisms and their evolutionary paths. Phylogenetic studies of several taxa measure similarities between defined characters. These characters can be morphological — shape of bones, leaves etc. — or molecular — the sequence of bases in DNA or proteins. Proteins and DNA aren’t preserved for more than about 10 000 years. Studies on fossil groups thus usually use morphological data.

The character data are put into a data matrix. In table 1, the species (letters) are in rows, the characters (numerals) in columns. The coding for character is given by the numerals in the matrix. “0”, “1”, “2” etc. for different character states, “?” is unknown and “-“ means missing data. The matrix can then be used for cladistic analysis, usually by software program. Cladistic analysis uses shared characters between organisms to work out common ancestors. Analyses use parsimony to find the fewest character changes between taxa. The hypothetical relationships can then be printed to a cladogram (fig. 1).
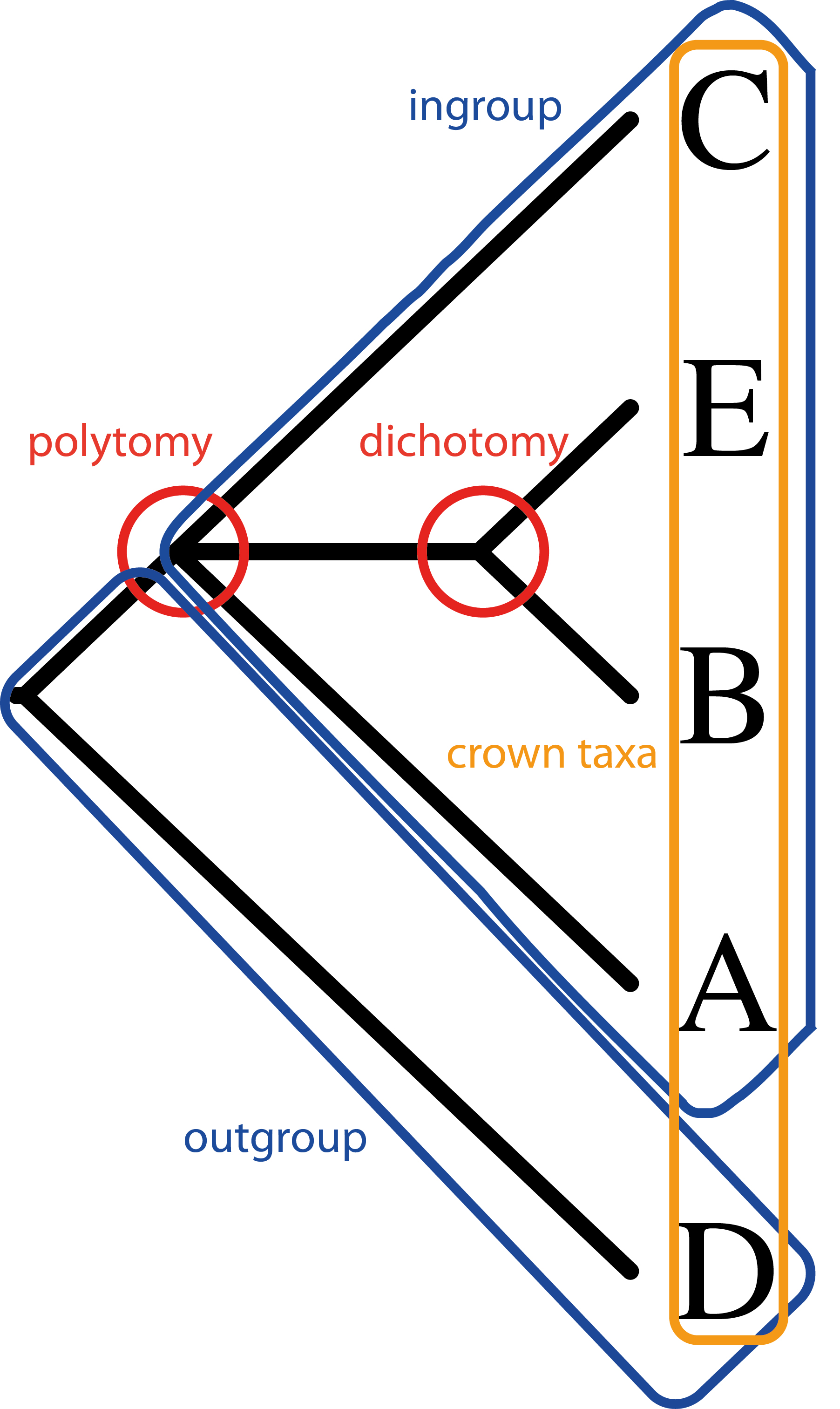
Cladograms show a hypothetical ‘family tree’ for the organisms in the phylogenetic study. Each branching shows a divergence from a common ancestor. Taxa on adjacent branches are more closely related than those separated by branches. Species on the lower branches are the outgroup whereas the inner branches contain the ingroup. Ingroup taxa contain a character that the outgroup taxa do not — a synapomorphy (apomorphy for a single taxon). A character shared by the ingroup and outgroup is a plesiomorphy. The same character evolving separately is called convergence or homoplasy.
Ichthyosaur phylogeny
At the turn of this millennium, three phylogenies of ichthyosaurs were published (Motani 1999; Maisch and Matzke 2000; Sander 2000). These were the first to look at the whole of the Ichthyosauria/Ichthyopterygia. Previous analyses had largely looked at Triassic taxa (e.g. Mazin 1982).
As these analyses were published so close to one another, each did not know of each other’s work. Thus, there are differences in the characters and codings used in these papers. Maisch and Matzke’s (2000) and Sander’s (2000) papers were published later enough to allow for comment on Motani’s (1999). Both of these compare their results to Motani’s. However, only Maisch and Matzke noted the numerous similarities between their characters and those of Motani’s. Maisch and Matzke were also able to incorporate characters from Motani that they did not have. As is to be expected from three independent analyses, the results differ in various places (fig. 2). Maisch (2010) discusses the main differences between them.
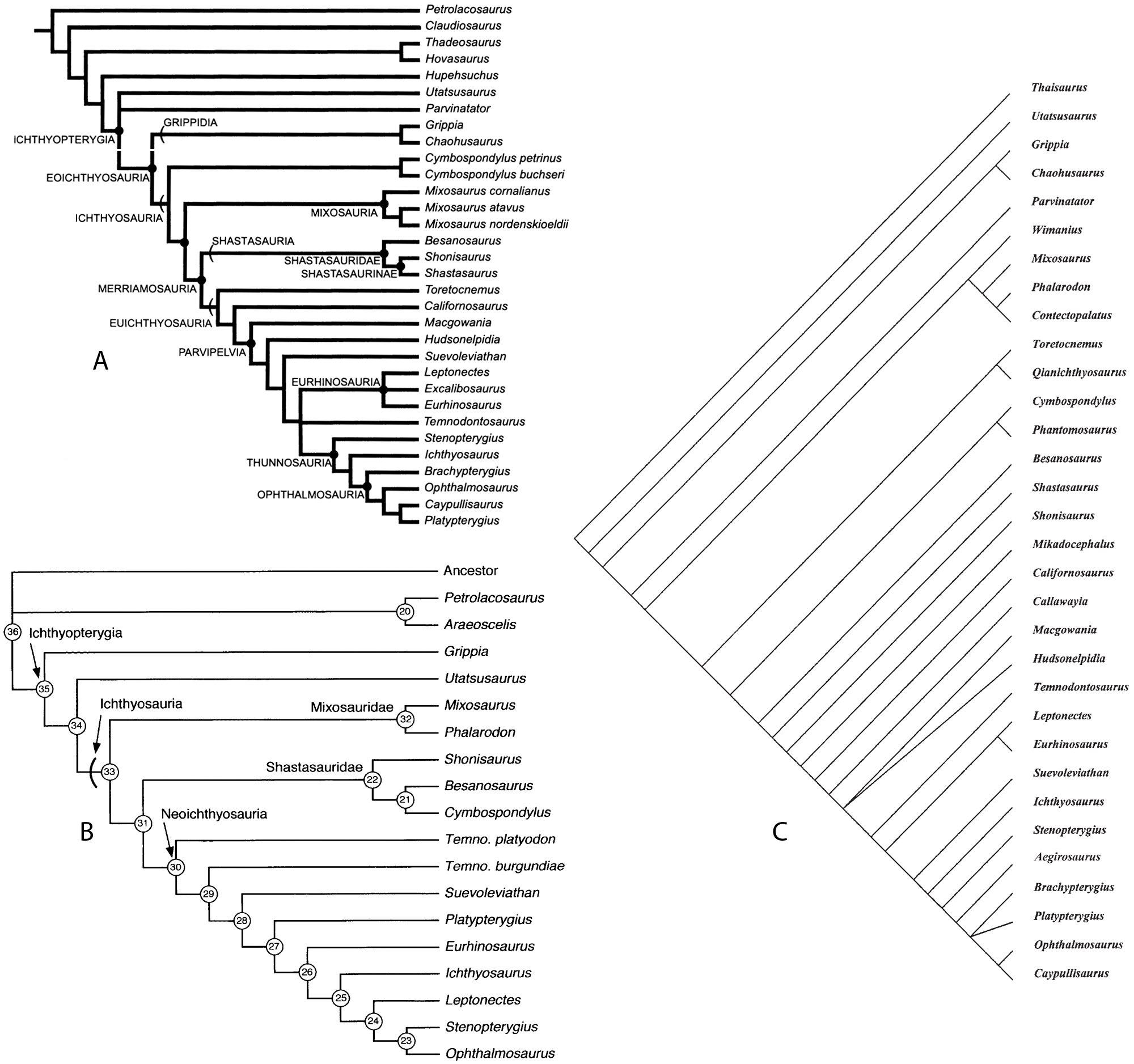
Subsequent descriptions of ichthyosaurs have often included a phylogenetic analysis. These are usually based upon Motani’s (1999) analysis, modified to accept new taxa and character modifications. The presence of three conflicting phylogenetic hypotheses is unsettling. A similar situation was found in the related Plesiosauria. These were another Mesozoic marine reptile group, including the four-flippered plesiosaurs and pliosaurs. This was recently discussed by Ketchum and Benson (2010).
Ketchum and Benson (2010) looked at two global scale and 11 small scale phylogenetic analyses of the Plesiosauria. Their critical analysis considered:
- Taxa used (66): increased number likely increases the accuracy of the phylogenetic hypothesis.
- Characters (178): all characters were re-examined based upon observation.
- Character coding: hierarchical (discrete or ‘conventional’) coding was deemed most appropriate; where continuous codings were used, gap-weighting (Thiele 1993) was use to ‘discretise’ the codings.
- Analysis: the Parsimony Ratchet system of Nixon (1999) was used to allow searching through the large matrix; heuristic and consensus searches were then completed using PAUP* Version 4.0β10 (Swofford 2002).
My plan is to use similar techniques upon all ichthyosaur cladistic analyses to date. The initial study will look to rectify the differences between Motani (1999), Maisch and Matzke (2000) and Sander (2000). These incorporate previous analyses, but I will look at pre-1999 studies to assess the characters that may have been removed subsequently. I shall also look at subsequent studies to assess modifications and the how my amalgamation of Motani, Maisch and Matzke and Sander affects them.
This concludes the second project within my thesis. Next time I will look at the further projects that will result from this, and the previous studies.
References
FELSENSTEIN, J. 2009. PHYLIP: Phylogeny inference package. University of Washington, USA. Downloaded from [http://evolution.gs.washington.edu/phylip.html] on 18 November 2009.
KETCHUM, H. F. and BENSON, R. B. J. 2010. Global interrelationships of Plesiosauria (Reptilia, Sauropterygia) and the pivotal role of taxon sampling in determining the outcome of phylogenetic analyses. Biological Reviews, 85, 361–392.
MAISCH, M. W. 2010. Phylogeny, systematics, and origin of the Ichthyosauria—the state of the art. Palaeodiversity, 3, 151–214.
MAISCH, M. W. and MATZKE, A. T. 2000. The Ichthyosauria. Stuttgarter Beiträge zur Naturkunde, Serie B (Geologie und Paläontologie), 298, 1–160.
MAZIN, J.-M. 1982. Affinités et phylogénie des Ichthyopterygia. *Geobios, **6, 85–98.
MOTANI, R. 1999. Phylogeny of the Ichthyopterygia. Journal of Vertebrate Paleontology, 19, 473–496.
NIXON, K. C. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics, 15, 407–414.
SANDER, P. M. 2000. Ichthyosauria: their diversity, distribution, and phylogeny. Paläontologische Zeitschrift, 74, 1–35.
SWOFFORD, D. 2002. PAUP*: Phylogenetic Analysis Using Parsimony. Sinauer Associates, Sunderland, Massachusetts, USA.
THIELE, K. 1993. The holy grail of the perfect character: the cladistic treatment of morphometric data. Cladistics, 9, 275–304.

Leave a comment